Maurice Wilkins, the nuclear physicist, entered the race for DNA based on a stroke of luck. After his work with the Manhattan Project on atomic bombs was completed, he wanted to switch to a more peaceful line of work and was inspired to investigate the physical basis for life. He turned to the fast-growing field of biophysics, taking up a position at the University of London. Early in his career there, he happened to attend a conference where a biochemist gave away samples of high-quality DNA. Wilkins was lucky enough to get a sample — though it might not have seemed that impressive at the time. It was so slimy and gooey that he later described it as “just like snot.” Nevertheless, because it contained long, intact DNA molecules — which were hard to come by at the time — this slippery sample would turn out to be critical in uncovering clues to DNA’s structure. Raymond Gosling, a Ph.D. student in Wilkins’ lab, suggested looking at the DNA with a new observational technique called X-ray diffraction.
X-ray diffraction, developed in the first half of the 20th century, was one of the new technologies that made solving the structure of DNA possible. The technique works on crystals, a kind of molecule with a regular, repeating structure. When X-rays are aimed through a sample, they are bent or diffracted in different directions depending on the locations of the atoms in the sample, and the final direction of the X-rays can be recorded on film. Because the X-rays must travel through many layers of atoms, it’s important that the atoms always occur in the same crystalline arrangement. If they don’t, the X-rays are bent into overlapping patterns, leaving the results a fuzzy, indistinct blur. However, if the structure has a repeating arrangement of atoms, they leave a pattern of sharp, clear spots. Different structures scatter the X-rays into different characteristic patterns.
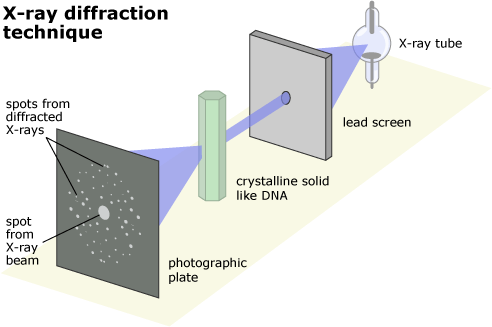
Even though scientists couldn’t directly observe the atoms within the crystal, they could work backward from X-ray diffraction patterns to reconstruct the three-dimensional structure that produced the scattering. This works a little like trying to figure out how tall a person is by looking at his or her shadow. Depending on the angle of the sun, the shadow might be longer or shorter, but if you could compare many pictures of their shadow at different times of day, you’d eventually be able to figure out how tall they were. Similarly, scientists compare many “shadows,” or X-ray diffraction patterns, cast by a crystal to determine the arrangement of atoms within it.
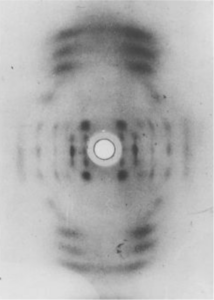
Although the DNA didn’t look very crystalline, Gosling wanted to try X-ray diffraction on the molecule anyway. Wilkins and Gosling knew DNA’s structure might be too irregular to produce a clear, well-defined X-ray pattern, but as it turned out, the sample was sticky and stringy because it was made up of lots of long, thin molecules of intact, crystalline DNA. Over the summer of 1950, Wilkins and Gosling’s patterns showed that DNA did have a regular structure — which meant that X-ray diffraction would be a critical tool in solving the structure. The patterns even suggested what that basic structure might be. Despite a few confusing blurry spots, the images hinted that DNA might come in the form of a twisted spiral — better known as a helix — though it was still not clear how the phosphates, sugars, and bases were arrayed within that helix.
DNA is much too small for us to observe its structure directly with our own eyes. Fortunately, scientists can often use tools (like X-ray diffraction) to extend and refine our basic senses. To learn more about this, visit Observation beyond our eyes.